You can see all features showcased in the demo notebook
nlp.viz(pipe,data)
Visualize input data with an already configured Spark NLP pipeline,
for Algorithms of type (Ner,Assertion, Relation, Resolution, Dependency)
using Spark NLP Display
Automatically infers applicable viz type and output columns to use for visualization.
Example:
# works with Pipeline, LightPipeline, PipelineModel,PretrainedPipeline List[Annotator]
ade_pipeline = PretrainedPipeline('explain_clinical_doc_ade', 'en', 'clinical/models')
text = """I have an allergic reaction to vancomycin.
My skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums.
I would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication."""
nlp.viz(ade_pipeline, text)
returns:
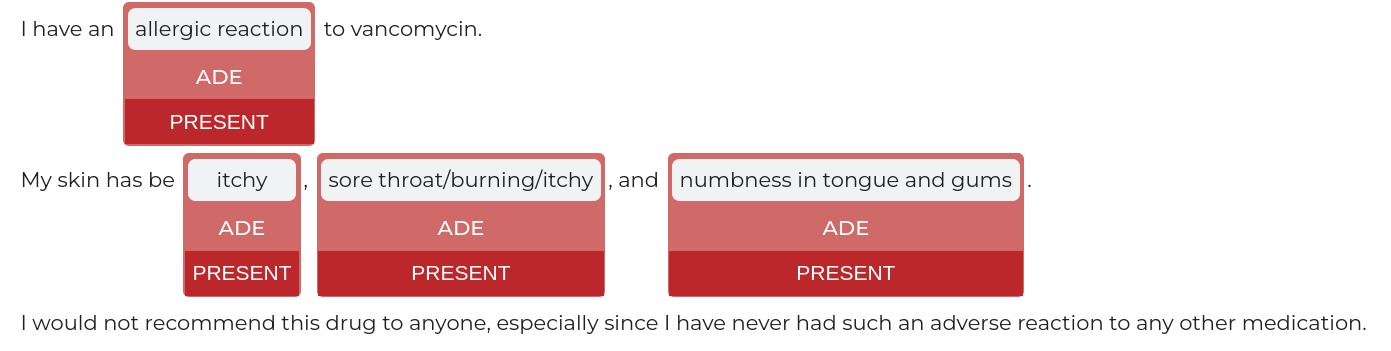
If a pipeline has multiple models candidates that can be used for a viz,
the first Annotator that is vizzable will be used to create viz.
You can specify which type of viz to create with the viz_type parameter
Output columns to use for the viz are automatically deducted from the pipeline, by using the
first annotator that provides the correct output type for a specific viz.
You can specify which columns to use for a viz by using the
corresponding ner_col, pos_col, dep_untyped_col, dep_typed_col, resolution_col, relation_col, assertion_col, parameters.
nlp.autocomplete_pipeline(pipe)
Auto-Complete a pipeline or single annotator into a runnable pipeline by harnessing NLU’s DAG Autocompletion algorithm and returns it as NLU pipeline.
The standard Spark pipeline is avaiable on the .vanilla_transformer_pipe attribute of the returned nlp pipe
Every Annotator and Pipeline of Annotators defines a DAG of tasks, with various dependencies that must be satisfied in topoligical order.
NLU enables the completion of an incomplete DAG by finding or creating a path between
the very first input node which is almost always is DocumentAssembler/MultiDocumentAssembler
and the very last node(s), which is given by the topoligical sorting the iterable annotators parameter.
Paths are created by resolving input features of annotators to the corrrosponding providers with matching storage references.
Example:
# Lets autocomplete the pipeline for a RelationExtractionModel, which as many input columns and sub-dependencies.
from sparknlp_jsl.annotator import RelationExtractionModel
re_model = RelationExtractionModel().pretrained("re_ade_clinical", "en", 'clinical/models').setOutputCol('relation')
text = """I have an allergic reaction to vancomycin.
My skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums.
I would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication."""
nlu_pipe = nlp.autocomplete_pipeline(re_model)
nlu_pipe.predict(text)
returns :
| relation | relation_confidence | relation_entity1 | relation_entity2 | relation_entity2_class |
|---|---|---|---|---|
| 1 | 1 | allergic reaction | vancomycin | Drug_Ingredient |
| 1 | 1 | skin | itchy | Symptom |
| 1 | 0.99998 | skin | sore throat/burning/itchy | Symptom |
| 1 | 0.956225 | skin | numbness | Symptom |
| 1 | 0.999092 | skin | tongue | External_body_part_or_region |
| 0 | 0.942927 | skin | gums | External_body_part_or_region |
| 1 | 0.806327 | itchy | sore throat/burning/itchy | Symptom |
| 1 | 0.526163 | itchy | numbness | Symptom |
| 1 | 0.999947 | itchy | tongue | External_body_part_or_region |
| 0 | 0.994618 | itchy | gums | External_body_part_or_region |
| 0 | 0.994162 | sore throat/burning/itchy | numbness | Symptom |
| 1 | 0.989304 | sore throat/burning/itchy | tongue | External_body_part_or_region |
| 0 | 0.999969 | sore throat/burning/itchy | gums | External_body_part_or_region |
| 1 | 1 | numbness | tongue | External_body_part_or_region |
| 1 | 1 | numbness | gums | External_body_part_or_region |
| 1 | 1 | tongue | gums | External_body_part_or_region |
nlu.to_pretty_df(pipe,data)
Annotates a Pandas Dataframe/Pandas Series/Numpy Array/Spark DataFrame/Python List strings /Python String
with given Spark NLP pipeline, which is assumed to be complete and runnable and returns it in a pythonic pandas dataframe format.
Example:
# works with Pipeline, LightPipeline, PipelineModel,PretrainedPipeline List[Annotator]
ade_pipeline = PretrainedPipeline('explain_clinical_doc_ade', 'en', 'clinical/models')
text = """I have an allergic reaction to vancomycin.
My skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums.
I would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication."""
# output is same as nlp.autocomplete_pipeline(re_model).nlp_pipe.predict(text)
nlp.to_pretty_df(ade_pipeline,text)
returns :
| assertion | asserted_entitiy | entitiy_class | assertion_confidence |
|---|---|---|---|
| present | allergic reaction | ADE | 0.998 |
| present | itchy | ADE | 0.8414 |
| present | sore throat/burning/itchy | ADE | 0.9019 |
| present | numbness in tongue and gums | ADE | 0.9991 |
Annotators are grouped internally by NLP into output levels token,sentence, document,chunk and relation
Same level annotators output columns are zipped and exploded together to create the final output df.
Additionally, most keys from the metadata dictionary in the result annotations will be collected and expanded into their own columns in the resulting Dataframe, with special handling for Annotators that encode multiple metadata fields inside of one, seperated by strings like ||| or :::.
Some columns are omitted from metadata to reduce total amount of output columns, these can be re-enabled by setting metadata=True
For a given pipeline output level is automatically set to the last annotators output level by default.
This can be changed by defining to_preddty_df(pipe,text,output_level='my_level' for levels token,sentence, document,chunk and relation .
nlp.to_nlu_pipe(pipe)
Convert a pipeline or list of annotators into a NLU pipeline making .predict() and .viz() avaiable for every Spark NLP pipeline.
Assumes the pipeline is already runnable.
# works with Pipeline, LightPipeline, PipelineModel,PretrainedPipeline List[Annotator]
ade_pipeline = PretrainedPipeline('explain_clinical_doc_ade', 'en', 'clinical/models')
text = """I have an allergic reaction to vancomycin.
My skin has be itchy, sore throat/burning/itchy, and numbness in tongue and gums.
I would not recommend this drug to anyone, especially since I have never had such an adverse reaction to any other medication."""
nlu_pipe = nlp.to_nlu_pipe(ade_pipeline)
# Same output as nlu.to_pretty_df(pipe,text)
nlu_pipe.predict(text)
# same output as nlu.viz(pipe,text)
nlu_pipe.viz(text)
# Acces auto-completed Spark NLP big data pipeline,
nlu_pipe.vanilla_transformer_pipe.transform(spark_df)
returns :
| assertion | asserted_entitiy | entitiy_class | assertion_confidence |
|---|---|---|---|
| present | allergic reaction | ADE | 0.998 |
| present | itchy | ADE | 0.8414 |
| present | sore throat/burning/itchy | ADE | 0.9019 |
| present | numbness in tongue and gums | ADE | 0.9991 |
and
